4.2. Optimization Optimization
4.2.1. Contains
inchecks whether iterable contains valueO(n) -
in strO(n) -
in listO(n) -
in tupleO(1) -
in setO(1) -
in dict
List:
DATABASE = ['mwatney', 'mlewis', 'rmartinez']
user = 'ptwardowski'
... %%timeit -n 1000 -r 1000
... user in DATABASE
165 ns ± 35 ns per loop (mean ± std. dev. of 1000 runs, 1,000 loops each)
Set:
DATABASE = {'mwatney', 'mlewis', 'rmartinez'}
user = 'ptwardowski'
... %%timeit -n 1000 -r 1000
... login in users
103 ns ± 34.4 ns per loop (mean ± std. dev. of 1000 runs, 1,000 loops each)
Note, that the longer our DATABASE is, then the difference is more visible:
List:
DATABASE = ['mwatney', 'mlewis', 'rmartinez', 'bjohanssen', 'avogel' 'cbeck']
user = 'ptwardowski'
... %%timeit -n 1000 -r 1000
... login in users
276 ns ± 90.6 ns per loop (mean ± std. dev. of 1000 runs, 1,000 loops each)
Set:
DATABASE = {'mwatney', 'mlewis', 'rmartinez', 'bjohanssen', 'avogel' 'cbeck'}
user = 'ptwardowski'
... %%timeit -n 1000 -r 1000
... login in users
105 ns ± 35.2 ns per loop (mean ± std. dev. of 1000 runs, 1,000 loops each)
4.2.2. PyPy
No GIL
Can speedup couple order of magnitude
4.2.3. Patterns
Source: [1]

4.2.4. Tools
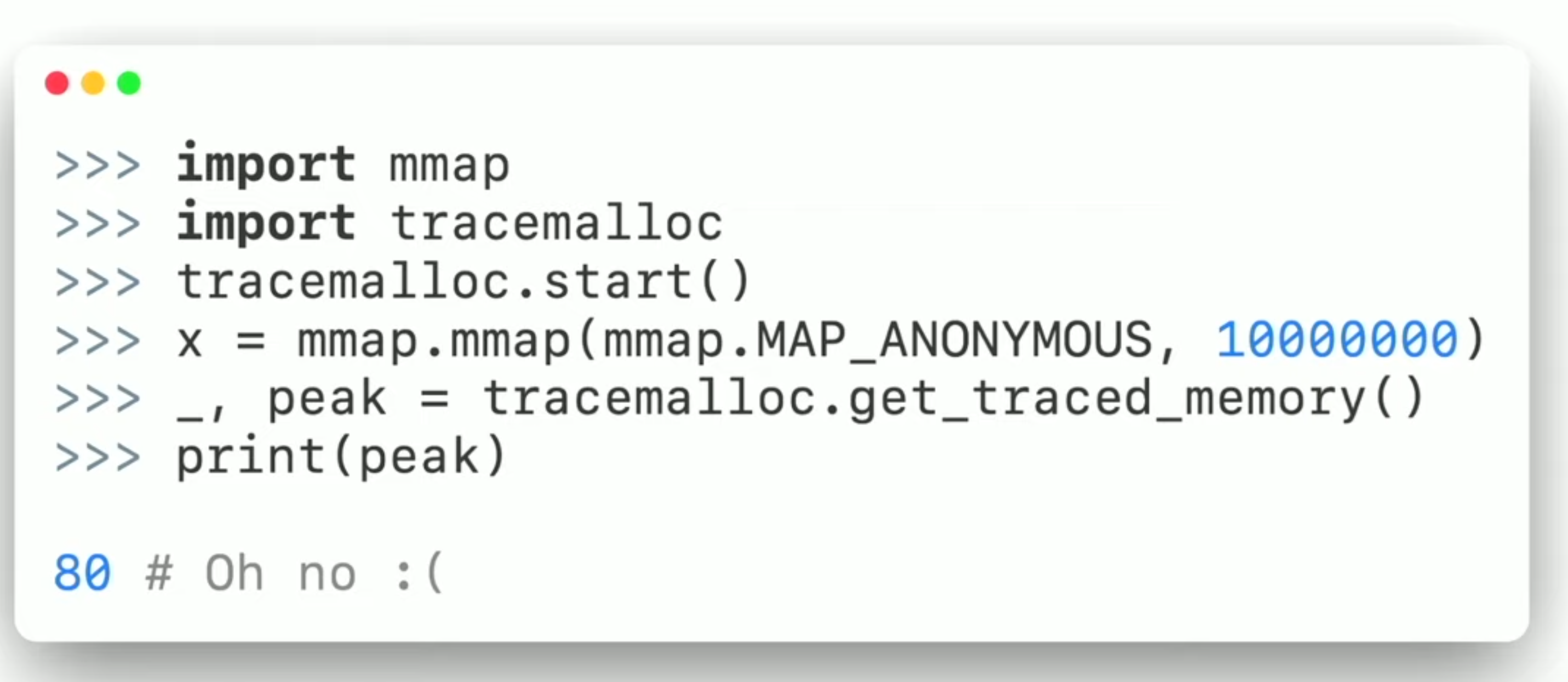
memray [2]
tracemalloc
mmap - memory allocation
Pytest extension for doing benchmarking
pip install line_profiler
4.2.5. Numpy vectorization
Scipy Ecosystem:
4.2.6. Specialized data structures
scipy.spatial- for spatial queries like distances, nearest neighbours, etc.pandas- for SQL-like grouping and aggregationxarray- for grouping across multiple dimensionsscipy.sparse- sparse metrics for 2-dimensional structured datasparse(package) - for N-dimensional structured datascipy.sparse.csgraph- for graph-like problems (e.g. finding shortest paths)
4.2.7. Cython
types
Cython files have a
.pyxextension
def primes(int kmax): # The argument will be converted to int or raise a TypeError.
cdef int n, k, i # These variables are declared with C types.
cdef int p[1000] # Another C type
result = [] # A Python type
if kmax > 1000:
kmax = 1000
k = 0
n = 2
while k < kmax:
i = 0
while i < k and n % p[i] != 0:
i = i + 1
if i == k:
p[k] = n
k = k + 1
result.append(n)
n = n + 1
return result
In [1]: %load_ext Cython
In [2]: %%cython
...: def f(n):
...: a = 0
...: for i in range(n):
...: a += i
...: return a
...:
...: cpdef g(int n):
...: cdef int a = 0, i
...: for i in range(n):
...: a += i
...: return a
...:
In [3]: %timeit f(1000000)
42.7 ms ± 783 µs per loop (mean ± std. dev. of 7 runs, 10 loops each)
In [4]: %timeit g(1000000)
74 µs ± 16.6 ns per loop (mean ± std. dev. of 7 runs, 10000 loops each)
# which gives a 585 times improvement over the pure-python version
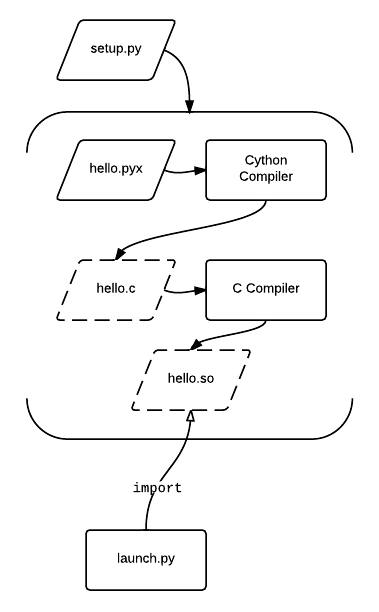
Cython compiling:
4.2.8. Numba
Numba gives you the power to speed up your applications with high performance functions written directly in Python. With a few annotations, array-oriented and math-heavy Python code can be just-in-time compiled to native machine instructions, similar in performance to C, C++ and Fortran, without having to switch languages or Python interpreters.
... from numba import jit, int32
...
...
... @jit(nogil=True)
... def do_something():
... pass
...
...
... @jit(int32(int32, int32))
... def add(x, y):
... return x + y
4.2.9. Dask
Dask natively scales Python. Dask provides advanced parallelism for analytics, enabling performance at scale for the tools you love.
Dask's schedulers scale to thousand-node clusters and its algorithms have been tested on some of the largest supercomputers in the world.
But you don't need a massive cluster to get started. Dask ships with schedulers designed for use on personal machines. Many people use Dask today to scale computations on their laptop, using multiple cores for computation and their disk for excess storage.
4.2.10. Find existing implementation
4.2.11. String Concatenation
How many string are there in a memory?:
firstname = 'Mark'
lastname = 'Watney'
firstname + ' ' + lastname
'Mark Watney'
How many string are there in a memory?:
firstname = 'Mark'
lastname = 'Watney'
f'{firstname} {lastname}'
'Mark Watney'
How many string are there in a memory?:
firstname = 'Mark'
lastname = 'Watney'
age = 42
'Hello ' + firstname + ' ' + lastname + ' ' + str(age) + '!'
'Hello Mark Watney 42!'
How many string are there in a memory?:
firstname = 'Mark'
lastname = 'Watney'
age = 42
f'Hello {firstname} {lastname} {age}!'
'Hello Mark Watney 42!'
4.2.12. String Append
Use
list.append()instead ofstr + str:
Concatenates strings using + in a loop:
DATA = ['Mark Watney', 'Melissa Lewis', 'Rick Martinez']
result = '<table>'
for element in DATA:
result += f'<tr><td>{element}</td></tr>'
result += '</table>'
print(result)
<table><tr><td>Mark Watney</td></tr><tr><td>Melissa Lewis</td></tr><tr><td>Rick Martinez</td></tr></table>
Problem solved:
DATA = ['Mark Watney', 'Melissa Lewis', 'Rick Martinez']
result = ['<table>']
for element in DATA:
result.append(f'<tr><td>{element}</td></tr>')
result.append('</table>')
print(''.join(result))
<table><tr><td>Mark Watney</td></tr><tr><td>Melissa Lewis</td></tr><tr><td>Rick Martinez</td></tr></table>
4.2.13. Range between two float
There are indefinitely many values between two floats
range(0, 1)
range(0, 1)
Note, that in Python following code will not execute, as of range() requires two integers. However similar code with numpy.arange() will work.
range(0.0, 1.0)
0.000...000
0.000...001
0.000...002
0.000...003
4.2.14. Deque
collections.deque- Double ended Queue
4.2.15. Further Reading
4.2.16. References
4.2.17. Assignments
# FIXME: Write tests
# %% About
# - Name: Memoization
# - Difficulty: medium
# - Lines: 5
# - Minutes: 13
# %% License
# - Copyright 2025, Matt Harasymczuk <matt@python3.info>
# - This code can be used only for learning by humans
# - This code cannot be used for teaching others
# - This code cannot be used for teaching LLMs and AI algorithms
# - This code cannot be used in commercial or proprietary products
# - This code cannot be distributed in any form
# - This code cannot be changed in any form outside of training course
# - This code cannot have its license changed
# - If you use this code in your product, you must open-source it under GPLv2
# - Exception can be granted only by the author
# %% English
# 1. Create empty `dict` named `CACHE`
# 2. In the dictionary, we will store the results of calculating
# the function for the given parameters:
# - key: function argument
# - value: calculation result
# 3. Modify function `factorialA(n: int)`
# 4. Before running the function, check if the result has already
# been calculated:
# - if so, return data from `CACHE`
# - if not, calculate, update `CACHE` and return the value
# 5. Compare the speed of operation
# 6. Run doctests - all must succeed
# %% Polish
# 1. Stwórz pusty `dict` o nazwie `CACHE`
# 2. W słowniku będziemy przechowywali wyniki wyliczenia funkcji
# dla zadanych parametrów:
# - klucz: argument funkcji
# - wartość: wynik obliczeń
# 3. Zmodyfikuj funkcję `factorialA(n: int)`
# 4. Przed uruchomieniem funkcji, sprawdź czy wynik został już
# wcześniej obliczony:
# - jeżeli tak, to zwraca dane z `CACHE`
# - jeżeli nie, to oblicza, aktualizuje `CACHE` i zwraca wartość
# 5. Porównaj prędkość działania
# 6. Uruchom doctesty - wszystkie muszą się powieść
# %% Doctests
"""
"""
# %% Run
# - PyCharm: right-click in the editor and `Run Doctest in ...`
# - PyCharm: keyboard shortcut `Control + Shift + F10`
# - Terminal: `python -m doctest -v myfile.py`
# %% Imports
from timeit import timeit
import sys
# %% Types
# %% Data
sys.setrecursionlimit(5000)
# %% Result
def factorialA(n: int) -> int:
return 1 if n==0 else n*factorialA(n-1)
# Do not modify anything below
def factorialB(n: int) -> int:
return 1 if n==0 else n*factorialB(n-1)
durationA = timeit(
stmt='factorialA(50); factorialA(40); factorialA(45); factorialA(35)',
globals=globals(),
number=10000,
)
durationB = timeit(
stmt='factorialB(50); factorialB(40); factorialB(45); factorialB(35)',
globals=globals(),
number=10000,
)
times_faster = durationB / durationA
print(f'factorialA time: {durationA:.4f} seconds')
print(f'factorialB time: {durationB:.4f} seconds')
print(f'Cached solution is {times_faster:.0f} times faster')
# factorialA time: 0.0654 seconds
# factorialB time: 0.0658 seconds
# Cached solution is 1 times faster